Who Am I?
I am a biomedical engineering PhD student at Boston University studying theoretical neuroscience under Dr. Brian DePasquale.
Research Interests
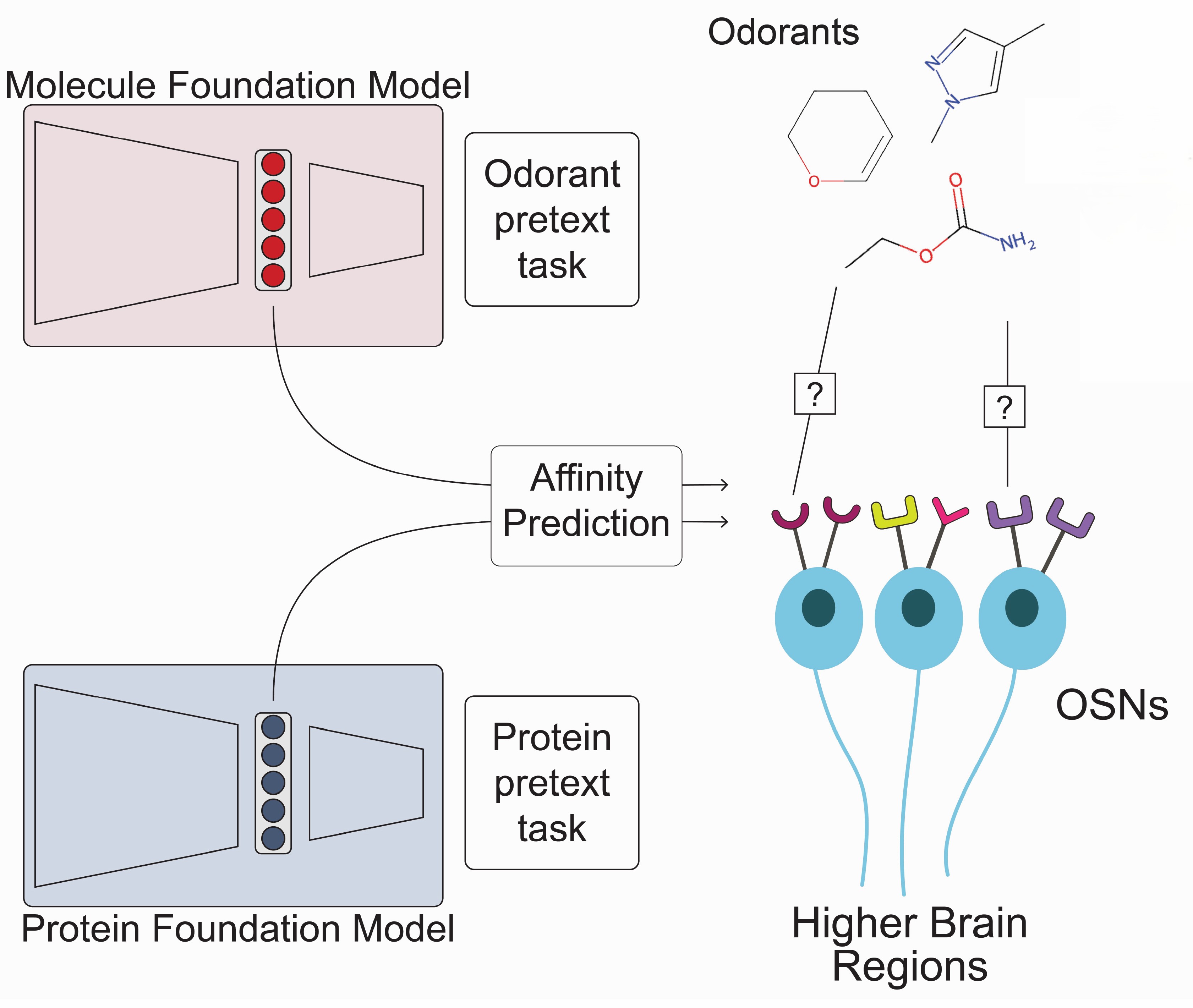
I’m interested in applying advanced neural network and machine learning models to study the brain and behavior. Currently, I’m working on two main projects. The first explores how odorous molecules interact with the mosquito olfactory system. To study this, I develop multimodal transformer models to predict odorant–olfactory receptor binding by integrating low-rank adapted protein and chemical foundation models. I’m also building custom chemical foundation models using graph neural networks (GNNs) specifically tailored for olfaction.
The second project focuses on using GNNs to study the collective behavior of schooling fish. In this work, I design GNNs that predict temporal, graph-structured data while simultaneously inferring dynamic edge relationships between individuals.
In my free time I like to make music, graphic design, and run.